Untargeted Lipidomics
Table of contents
Background
- Untargeted Lipidomics aims at the detection and relative quantification of membrane (structural) lipids. This covers phospholipids, sphingolipids, glycerolipids and cholesterol esters. The methods should also cover free fatty acids and acyl carnitines.
- Although all considerations are taken to perform an unbiased analysis, this does not mean that every lipid in those classes is detected. That is due to considerations of abundance, sensitivity, ionization efficiency and other factors. An example coverage data set for specific sample types (matrices) is attached below.
- It is important to consider that "untargeted" does not mean question-free experiments. Knowledge will not emerge from the mere collection of lot of data without clear questions defined apriori. Data torture after analysis performed to extract any knowledge without a question will mostly lead to finding confirming inherent biases and will mislead the interpretation. Therefore, it is crucial to spend the necessary time to reflect on the research question and how metabolomics will help answer it as this will be the topic of our first meeting.
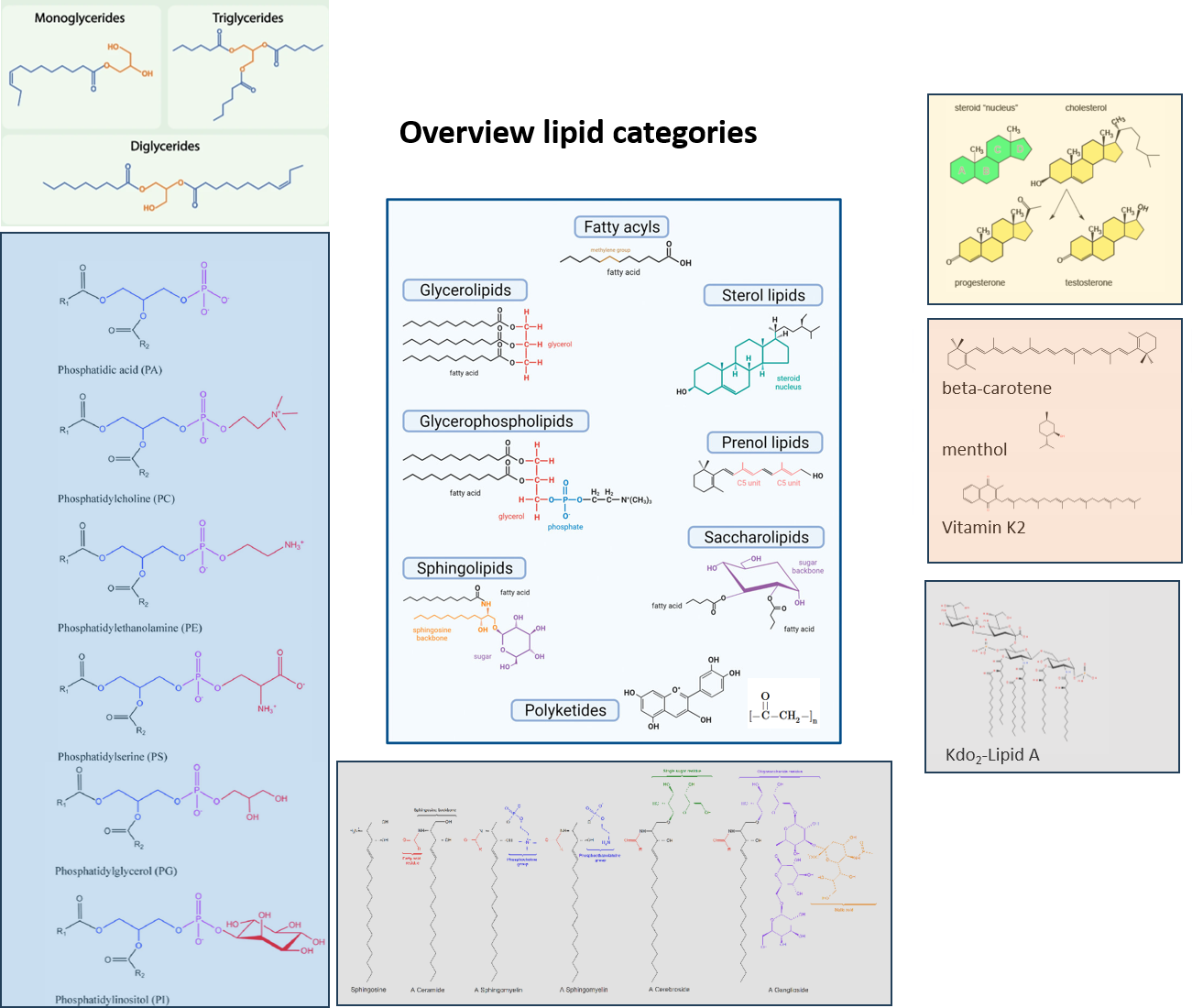
Lipid Classes overview
- Workflow :
- Study Design: It is important to consider a design that allows to answer the research question based on our current best knowledge. Check the literature, formulate a clear research question, try to mentally simulate the best (and worst) possible outcomes.
- Sample Collection: As metabolism is very fast and continuous process, extra care should be taken to stop metabolism (quenching) once the system is out of its natural state. Different protocols might be applied for quenching.
- Sample Preparation: As described below, this is usually done in our labs at FGCZ.
- LC/MS measurement: several analytical workflows exist to cover different chemical classes as described below in detail
- Data Analysis: This entails the analysis of raw mass spectrometry files for peak picking, alignment, normalization etc. as described below.
- Interpretation: This steps answers the research question through the acquired metabolomics datasets and ensuing analysis.
- New questions with new study design: As the process is iterative, usually successful experiments will lead to more targeted questions and hypothesis that can be refined by follow the same workflow or performing targeted LC-MS.
- References
Sample Preparation
Tissue Homogenization
Equipment needed
- TissueLyserII (or equivalent)
- Vortexer
- Eppendorf Thermomixer
- Eppendorf tubes (PCR clean)
- Temperature-controlled centrifuge
- SpeedVac or Nitrogen evaporator
- Stainless still or zirconia beads
- Scalpels for dissection
- Forceps
- Pipettes
Buffers and solvents
- MilliQ Water
- 1-butanol/methanol (1:1, v/v) with 5 mM ammonium formate
- methanol: isopropanol 1:1 (v/v)
Procedure: Tissue dissection
- Label and weigh the pre-cooled ( +4°C) Eppendorf tubes containing the beads and record the weight
- Cut a small piece of frozen tissue (approx. 10-20 mg) in the labeled tube or transfer suitable amount of microorganism. For lipid-rich organs such as the liver, brain, nerve, and adipose tissue 1-5 mg is enough
- Use surgical blades and scalpels. Use one surgical blade per group i.e. do not use the same blades for different groups. If the same blade has to be used for all samples, rinse it with ethanol and dry it between the groups.
- Store at -20 till homogenization
IMPORTANT: For organs with different distinct anatomic regions cut a slice from the same anatomic region in all samples (e.g. frontal lobe or cerebellum in the brain etc.) as different regions will have different lipidomics profiles and note the area for the protocol.
IMPORTANT: Do not let the tissues completely thaw in the process of the slicing, It is preferable to slice a few samples at a time and always keep the tissue samples on ice or a cooled metal rack when they are not in the freezer.
IMPORTANT: for organs that are rich in blood (e.g. liver), proper removal of blood through perfusion during animal sacrifice is recommended depending on the sacrifice method. If that is not possible, the lipidomics profile will reflect partly the blood content of that organ.
Procedure: Homogenization
- Add 500-1000 µl of pre-cooled ( -20°C) extraction buffer (water) to each sample containing tube. As a general rule of thumb, the volumetric ratio of the beads:tissue:water should be 1:1:1. Use the same amount of water to biological sample will result in a normalization of the input.
- Use TissueLyserII for tissue homogenization and cool the tubes in between the procedure on an ice bath ( +4°C). Perform a 2 to 5 min homogenization at 30Hz
- Remove the beads (Optional: recover for cleaning)
- Inspect the homogenate, no streaks of tissue should be left intact ( especially important for nerve and muscle tissues)
- Freeze the homogenate
Further processing
- Centrifuge homogenates for 10min at 10`000g (pre-cooled centrifuge, 4°C)
- Transfer all the supernatant to clean Eppendorf tube (Safe Lock)
- If the homogenization was performed in water, proceed with metabolites extraction in the buffer of choice. (=> Single Phase Extraction, Liquid-Liquid Extraction, Solid Phase Extraction)
- Keep the pellet and use it for protein quantification (all extraction buffer should be evaporated for correct protein quantification)
Single Phase Extraction
Lipid extraction from tissue homogenate or biofluids with BUME
- Add 100 μL of 1-butanol/methanol (1:1, v/v) with 5 mM ammonium formate to 10 μL of the biofluid or tissue homogenate
- Vortex the samples for 30 sec
- Incubate 30 minutes shaker 20°C 1000 rpm (alternative: sonicate in water batch 60 minutes, 20°C)
- Centrifuge the samples for 10 minutes 16`000g / 20°C
- Transfer a fixed amount of the supernatant to clean Eppendorf tubes for drying. Avoid touching the pellet (used for protein quantification). Alternative: use the extract directly for LC-MS
- The supernatant can be stored at -80°C if drying is not possible
- Dry under N2 stream at 30°C (ca. 10 minutes required)
- reconstitute dried sample in methanol:isopropanol 1:1 (v/v)
- inject 5- 10 μL on LC
Extraction Protocol for LC/MS lipidomics of Adherent cells
Lipidomics single phase methanol: isopropanol 1:1 (v/v) mixture addition to cell-culture wells.- Remove media gently from the cell culture wells
- Wash 1x with PBS ( volume equal to media removed)
- Remove the PBS after 10 seconds
- Add 1000 μL of cold (-20°C) methanol: isopropanol 1:1 (v/v) mixture to every well
- Manually shake the plate for 30 seconds
- Incubate the plates with closed lids at -80°C (5 min).
- Inspect the wells under the microscope to ensure that complete detachment of the cells has occurred. If not, use a scraper to detach the cells
- Make sure that complete cell lysis and detachment happens by checking the wells under the microscope after removal of the solvent
- Transfer the detached cells and solvent to Eppendorf tubes (or deep-well plates) and store them at -80°C.
- Once all samples are ready, take them out of the -80 and centrifuge them for 10 minutes at 2000g (for DWP plates) 16000g (for tubes).
- Collect the supernatant and dry it in the speed Vac (around 2 hours) or under N2
- Re-suspend the samples in 50 μl MeOH: IPA (1:1) by vortexing and mixing in a thermomixer for 10 minutes
- Centrifuge for 10 minutes at 16000g
- Transfer the supernatant to glass LC/MS vials or 96-well plates
- Solvents should be of the highest purity available (usually > 99.9%)
- Since it is not avoidable to use plastic tubes, it is recommended to use the highest quality plastic (e.g., Eppendorf safe lock tubes and not the Nunc plastic products)
- Different plastic types will lead to other background ions, so all samples should be extracted in the same plastic tubes
- The volume of extraction solvent per well has to be adjusted to cover the whole surface area of the well without a significant loss in evaporation
- For most applications, the number of cells needed ranges between 1e5-1e6. The more is, the better
- For normalization to cell number or protein content, approx. Cell number per well or protein content is needed ( e.g., from a parallel experiment)
Liquid-Liquid Extraction (LLE)
method adapted from:- Matyash et al 2008 https://doi.org/10.1194/jlr.D700041-JLR200
- SIMPLEX protocol from Coman et al 2016 10.1074/mcp.M115.053702
- Add 150 μL methanol to 20 μL of sample (e.g. biofluid or tissue homogenate). (volumetric ratio 7.5:1 methanol:sample)
- Vortex the samples for 30 sec
- Add 500 μl of MTBE to the mix. (volumetric ratio 3.3:1 MTBE:methanol)
- Incubate the mixture for 1 h at 25°C in a shaker.
- Add 125 μl of water.
- Centrifuge at 10`000 g for 10 min, 25°C
- Collect the upper (organic) phase for lipidomics. Approx. 450 μl
- Dry under nitrogen or using SpeedVac
- reconstitute dried sample in methanol: isopropanol 1:1 (v/v)
- inject 5- 10 μL on LC
Liquid Chromatography
C18 RPLC (Lipid mediators Gradient)
- Method name: FIAplus (pos, neg)
- Goal: untarget lipid mediators on C18, covering polar lipids, steroids, high throughput
- tested on: plasma, urine
LC method
- LC Column: Waters Acquity Premier BEH C18 1.7 µm 50mm x 2.1mm
- Column Temperature: 50°C
- Flow Rate: 1ml/min
- Run time: 5 min
- Eluents:
- A: H2O, 0.1% FA
- B: MeOH, 0.1% FA
- Gradient:
| Time (min) | Flow (ml/min) | % A | % B | Curve | |
| 1 | 0 | 1 | 90 | 10 | 5 |
| 2 | 0.47 | 1 | 90 | 10 | 5 |
| 3 | 3.07 | 1 | 0 | 100 | 5 |
| 4 | 4.07 | 1 | 0 | 100 | 5 |
| 5 | 4.27 | 1 | 90 | 10 | 5 |
| 6 | 4.87 | 1 | 90 | 10 | 5 |
- Injection volume: 5 to 10 ul
- Needle Wash: isopropanol: MeOH:ACN:H2O (1:1:1:1:1), 2% FA
- Sample solvent: H2O:MeOH (1:1)
- Autosampler Temperature: 20°C
- Sample vial/plates: Total recovery glass vials or 96 well plates
C18 RPLC (Lipidomics Gradient)
- Method name: Lipidomics (pos, neg)
- Goal: untarget lipidomics on C18
- tested on: Heart, Liver, Kidney, Adipose tissue, C. elegance, Myeloid cells, plasma, urine
LC method
- LC Column: Waters Acquity Premier BEH C18 1.7 µm 50mm x 2.1mm
- Column Temperature: 60°C
- Flow Rate: 1ml/min
- Run time: 7.5 min
- Eluents:
- A: ACN: H2O (60:40), 5 mM NH4 acetate
- B: Isopropanol: ACN (90:10), 5 mM NH4 acetate
- Gradient:
| Time (min) | Flow (ml/min) | % A | % B | Curve | |
| 1 | 0 | 1 | 85 | 15 | 5 |
| 2 | 1 | 1 | 70 | 30 | 5 |
| 3 | 1.25 | 1 | 52 | 48 | 5 |
| 4 | 5.5 | 1 | 18 | 82 | 5 |
| 5 | 5.75 | 1 | 1 | 99 | 5 |
| 6 | 6 | 1 | 1 | 99 | 5 |
| 7 | 6.05 | 1 | 1 | 99 | 5 |
| 8 | 7.5 | 1 | 70 | 30 | 5 |
- Injection volume: 5 to 10 ul
- Needle Wash: isopropanol:MeOH:ACN:H2O (1:1:1:1:1), 2% FA
- Sample solvent: Isopropanol:MeOH (1:1) or BUME (1:1)
- Autosampler Temperature: 20°C
- Sample vial/plates: Total recovery glass vials or 96 well plates
- Method file:
- pos: sstreb_20230526_lipidomics_DDA5_pos_1ml_NCE10_20_30_mz_200_2000_8min
- neg: sstreb_20230526_lipidomics_DDA5_neg_1ml_NCE10_20_30_mz_200_2000_8min
- Tune file: HESI_20230208_Lipidomics_AO
Mass Spectrometry
Ionization Parameters
- Polarity: HESI negative (3.5kV), HESI positive (3.8kV)
MS1 parameters
- Microscans: 1
- Resolution: 60,000
- AGC target: 3e6
- Maximum IT: 100 ms
- Number of scan ranges: 1
- Scan range: 200 to 2000 m/z
- Spectrum data type: Profile
MS2 parameters, dd-MS2
- Microscans: 1
- Resolution: 15,000
- AGC target: 1e5
- Maximum IT: 50 ms
- Loop count: 5
- MSX count: 1
- TopN: 5
- Isolation window: 1.0 m/z
- Isolation offset: 0.0 m/z
- Fixed first mass: ―
- (N)CE / stepped (N)CE: 10, 20, 30
- Spectrum data type: Profile
- Minimum AGC target: 8e3
Data Analysis
Skyline
Skyline is a freely available, open-source Windows client application for building Selected Reaction Monitoring (SRM) / Multiple Reaction Monitoring (MRM), Parallel Reaction Monitoring (PRM), DIA/SWATH and targeted DDA quantitative methods and analyzing the resulting mass spectrometer data.Within the untargeted metabolomics workflow, it is used to monitor the quality of the acquired data during the acquisition process. A predefined set of lipids/metabolites, present in our in-house developed standard mixture, are regularly monitored during the acquisition of a batch of samples, both in the above-mentioned mixture of standards and in QC samples, to gain insights on system performance.
The transition list used in the QC process is available No such attachment on this page.
Compound Discoverer
Compound Discoverer is the software in use for pure untargeted data analysis. When the project focus is group differences investigation and features discovery. The output will be a relative difference ratio. Compound Discovery allows for dataset exploration in a modular and customizable fashion.The picture represents the Untargeted Lipidomics Workflow Tree generally used, each node comprises several parameters that can be manipulated depending on the dataset and the scope of the analysis.
- MS2 spectra libraries: lipidblast, mzcloud, NIST
Further details regarding the parameters setting used in the standard untargeted metabolomics pipeline can be found No such attachment on this page.
Method Validation
Expected Data
- Raw files are made available in the "Workunits" section of each B-Fabric order page
- Results are delivered in the form of Excel tables and a Result report, summarizing sample processing, data acquisition and analysis, and main findings.
typical figures provided to the users to evaluate their data
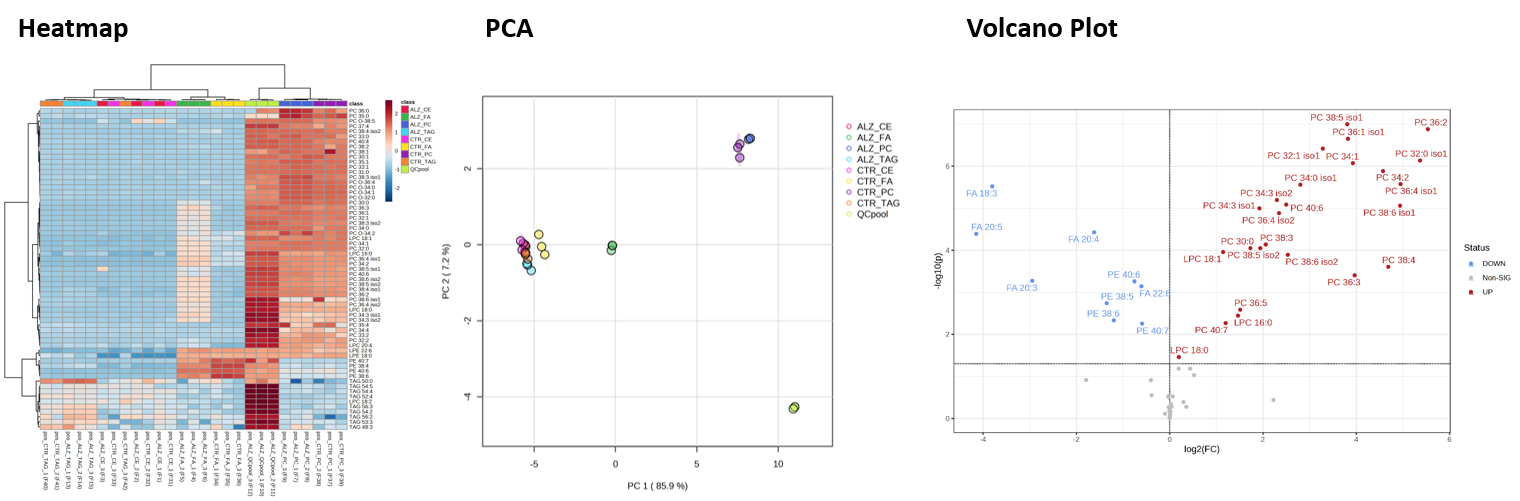
- on request we can provide csv files which can be directly used in metaboanalyst metaboanalyst, which allows statistical data analysis and visualization.